Gene editing technology is getting better and growing faster than ever before. New and improved base editors—an especially efficient and precise kind of genetic corrector—inch the tech closer to treating genetic diseases in humans.
copyright by phys.org
 But, the base editor boom comes with a new challenge: Like a massive key ring with no guide, scientists can sink huge amounts of time into searching for the best tool to solve genetic malfunctions like those that cause sickle cell anemia or progeria (a rapid aging disease). For patients, time is too important to waste.
But, the base editor boom comes with a new challenge: Like a massive key ring with no guide, scientists can sink huge amounts of time into searching for the best tool to solve genetic malfunctions like those that cause sickle cell anemia or progeria (a rapid aging disease). For patients, time is too important to waste.
“New base editors come out seemingly every week,” said David Liu, Thomas Dudley Cabot Professor of the Natural Sciences and a core institute member of the Broad Institute and the Howard Hughes Medical Institute (HHMI). “The progress is terrific, but it leaves researchers with a bewildering array of choices for what base editor to use.”
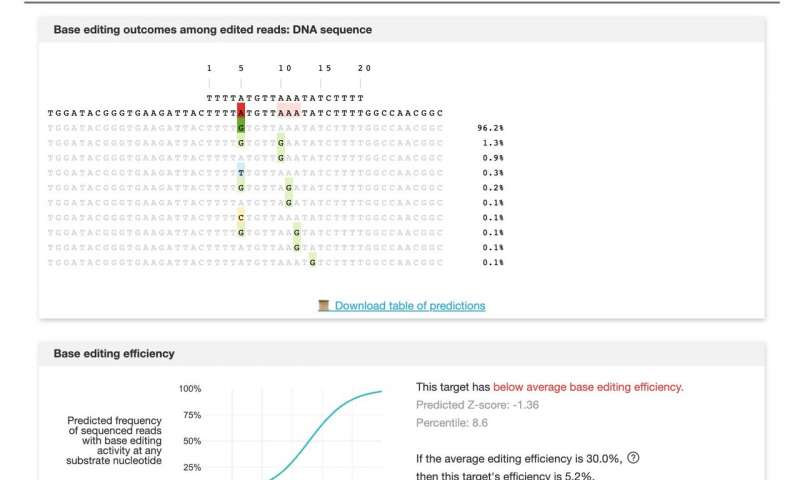
Liu invented base editors. Fittingly, he and his research team have now invented a way to identify which are most likely to achieve desired edits, as reported today in Cell . Using experimental data from editing more than 38,000 target sites in human and mouse cells with 11 of the most popular base editors (BEs), they created a machine learning model that accurately predicts base editing outcomes, Liu said. The library, called BE-Hive, is available for public use . But the effort produced more than a neat catalog of BEs; the machine learning model discovered new editor properties and capabilities that humans failed to notice.
“If you set out to use base editing to correct a single disease-causing mutation,” said Mandana Arbab, a postdoctoral fellow in the Liu lab and co-first author on the study, “you’re left with a mountain of possible ways to do it and it is difficult to know which ones are most likely to work.”
Base editors may be more precise than other forms of gene editing, but they can still cause unwanted, often unpredictable, edits outside the intended genetic target. Each editor has its own eccentricities. Different types operate within smaller or larger editing “windows,” stretches of DNA about two to five letters wide. Some editors might overshoot or undershoot their targets; others might change just one of two As in a given window.
“If the sequence within the window is GACA,” Liu said, “and you’re using an adenine base editor to change one of those As, will one be preferentially edited over the other?”
The answer depends on the base editor, its paired guide RNA—the chaperone that ferries the editor to the appropriate DNA work site—and the surrounding DNA sequence. To corral all these complicating factors, the team first collected a massive amount of data.[…]
Thank you for reading this post, don't forget to subscribe to our AI NAVIGATOR!
read more – copyright by phys.org

Gene editing technology is getting better and growing faster than ever before. New and improved base editors—an especially efficient and precise kind of genetic corrector—inch the tech closer to treating genetic diseases in humans.
copyright by phys.org
“New base editors come out seemingly every week,” said David Liu, Thomas Dudley Cabot Professor of the Natural Sciences and a core institute member of the Broad Institute and the Howard Hughes Medical Institute (HHMI). “The progress is terrific, but it leaves researchers with a bewildering array of choices for what base editor to use.”
Liu invented base editors. Fittingly, he and his research team have now invented a way to identify which are most likely to achieve desired edits, as reported today in Cell . Using experimental data from editing more than 38,000 target sites in human and mouse cells with 11 of the most popular base editors (BEs), they created a machine learning model that accurately predicts base editing outcomes, Liu said. The library, called BE-Hive, is available for public use . But the effort produced more than a neat catalog of BEs; the machine learning model discovered new editor properties and capabilities that humans failed to notice.
“If you set out to use base editing to correct a single disease-causing mutation,” said Mandana Arbab, a postdoctoral fellow in the Liu lab and co-first author on the study, “you’re left with a mountain of possible ways to do it and it is difficult to know which ones are most likely to work.”
Base editors may be more precise than other forms of gene editing, but they can still cause unwanted, often unpredictable, edits outside the intended genetic target. Each editor has its own eccentricities. Different types operate within smaller or larger editing “windows,” stretches of DNA about two to five letters wide. Some editors might overshoot or undershoot their targets; others might change just one of two As in a given window.
“If the sequence within the window is GACA,” Liu said, “and you’re using an adenine base editor to change one of those As, will one be preferentially edited over the other?”
The answer depends on the base editor, its paired guide RNA—the chaperone that ferries the editor to the appropriate DNA work site—and the surrounding DNA sequence. To corral all these complicating factors, the team first collected a massive amount of data.[…]
Thank you for reading this post, don't forget to subscribe to our AI NAVIGATOR!
read more – copyright by phys.org
Share this: