Insilico Medicine, a Rockville-based next-generation artificial intelligence company specializing in the application of deep learning for target identification, drug discovery and aging research announces the publication of a new research paper ” Machine learning on human muscle transcriptomic data for biomarker discovery and tissue-specific drug target identification ” in Frontiers in Genetics journal.
copyright by eurekalert.org
 Sarcopenia (from Greek “flesh poverty”), is one of the major age-related processes and involves the loss of skeletal muscle and its function. Age-associated muscle wasting remains an important clinical challenge that impacts hundreds of millions of older adults. It is associated with serious negative health outcomes such as falls, impaired standing balance, physical disability, and mortality. The many insights into sarcopenia from aging research suggest that understanding the molecular mechanisms of muscle aging can reveal novel potentially rejuvenating treatments.
Sarcopenia (from Greek “flesh poverty”), is one of the major age-related processes and involves the loss of skeletal muscle and its function. Age-associated muscle wasting remains an important clinical challenge that impacts hundreds of millions of older adults. It is associated with serious negative health outcomes such as falls, impaired standing balance, physical disability, and mortality. The many insights into sarcopenia from aging research suggest that understanding the molecular mechanisms of muscle aging can reveal novel potentially rejuvenating treatments.
To address this challenge researchers from Insilico Medicine developed a novel deep-learning based model that predicts a biological age of a muscle and can be used to estimate the relevant importance of the genetic and epigenetic factors driving this process within many age groups. The paper explaining one of the simple models for applying the age predictors developed using several machine learning techniques was published in Frontiers in Genetics .
“We are working on multiple biomarkers using deep learning and including blood biochemistry, transcriptomics, and even imaging data to be able to track the effectiveness of the various interventions we are developing. We believe that the most effective anti-aging therapy should be tissue-specific, so we focused on the development of tissue-specific biomarkers of aging. This work is an example of a marker of skeletal muscle tissue. Internally, we work with six tissues, including the liver, skin, and lungs.” said Polina Mamoshina, senior deep learning scientist at Insilico Medicine.
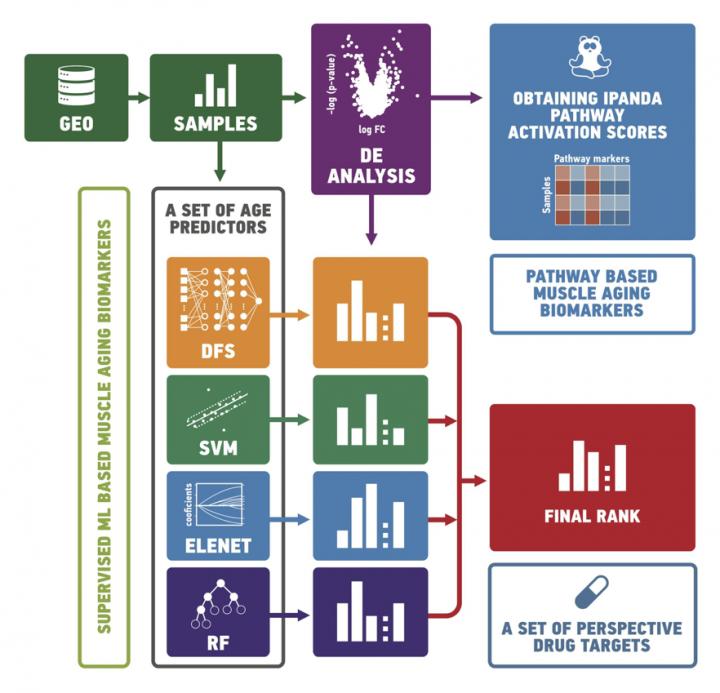
In this study, the scientists applied a state of the art signaling pathway analysis algorithm, iPANDA, to compare transcriptomic signatures of ‘old’ and ‘young’ tissues and utilized several machine learning methods to predict the age of samples based on their transcriptomic signatures. Ultimately, the trained age predictors were used to identify tissue-specific aging clocks.
This combined data-driven approach demonstrates that age prediction models can become a powerful tool for identifying prospective targets for geroprotectors. […]
read more – copyright by eurekalert.org

Insilico Medicine, a Rockville-based next-generation artificial intelligence company specializing in the application of deep learning for target identification, drug discovery and aging research announces the publication of a new research paper ” Machine learning on human muscle transcriptomic data for biomarker discovery and tissue-specific drug target identification ” in Frontiers in Genetics journal.
copyright by eurekalert.org
To address this challenge researchers from Insilico Medicine developed a novel deep-learning based model that predicts a biological age of a muscle and can be used to estimate the relevant importance of the genetic and epigenetic factors driving this process within many age groups. The paper explaining one of the simple models for applying the age predictors developed using several machine learning techniques was published in Frontiers in Genetics .
“We are working on multiple biomarkers using deep learning and including blood biochemistry, transcriptomics, and even imaging data to be able to track the effectiveness of the various interventions we are developing. We believe that the most effective anti-aging therapy should be tissue-specific, so we focused on the development of tissue-specific biomarkers of aging. This work is an example of a marker of skeletal muscle tissue. Internally, we work with six tissues, including the liver, skin, and lungs.” said Polina Mamoshina, senior deep learning scientist at Insilico Medicine.
In this study, the scientists applied a state of the art signaling pathway analysis algorithm, iPANDA, to compare transcriptomic signatures of ‘old’ and ‘young’ tissues and utilized several machine learning methods to predict the age of samples based on their transcriptomic signatures. Ultimately, the trained age predictors were used to identify tissue-specific aging clocks.
This combined data-driven approach demonstrates that age prediction models can become a powerful tool for identifying prospective targets for geroprotectors. […]
read more – copyright by eurekalert.org
Share this: